Introduction
Common bean (Phaseolus vulgaris L.) is an important food legume and is a main source of protein, hence its nickname ‘poor man’s meat’ (Broughton et al. 2003). It plays a vital role in agriculture by associating with rhizobia and fixing atmospheric N2 through a biological nitrogen fixation (BNF) process. A wide range of rhizobia is known to associate with beans due to its promiscuous nature and these different strains are likely to vary in their BNF potential. Thus, with the aim of identifying more effective rhizobia, we trapped new strains from bean growing areas in Southern Ethiopia and genotyped them to determine their taxonomic identity and relatedness to reference strains of known N2 fixing ability.
|
Materials and Methods Results and Discussion Conclusions |
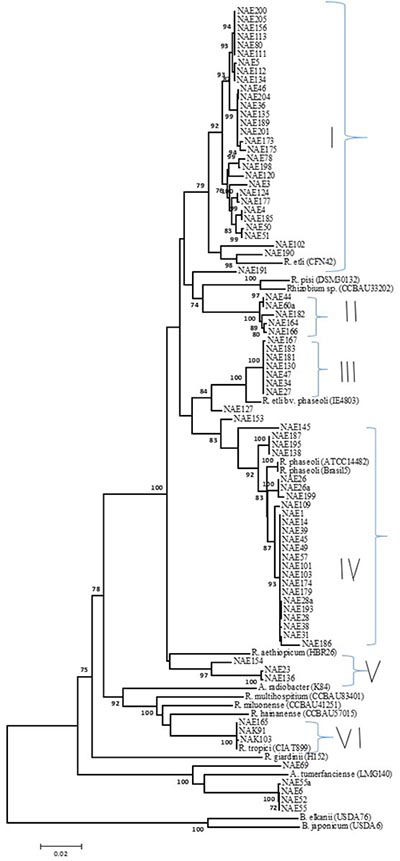
Figure 1: Neighbour-joining phylogeny of MLSA |
Ashenafi Hailu Gunnabo, Wageningen University & Research, The Netherlands (Click here for his 2017 update)
References
- 10.1016/j.syapm.2011.11.005
- Broughton WJ, Hernandez G, Blair M, et al (2003) Beans (Phaseolus spp.) - model food legumes W. Plant Soil 252:55–128. doi: 10.1023/A:1024146710611
- Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Evol Genet Anal 33:1870–1874. doi: 10.1093/molbev/msw054
- Mwenda GM, O’Hara GW, De Meyer SE, et al (2018) Genetic diversity and symbiotic effectiveness of Phaseolus vulgaris -nodulating rhizobia in Kenya. Syst Appl Microbiol. doi: 10.1016/j.syapm.2018.02.001
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. doi: 10.1093/nar/22.22.4673